Medical Image Segmentation
Medical image segmentation with UNet architectures
This study focuses on the medical image segmentation of CT and MRI scans and serves as a course project for IFT 3710/6759 H23 - Projets (avancés) en apprentissage automatique. It was conducted in collaboration with my classmates, Arthur Boschet and Frederic Gagne.
Medical image segmentation is a critical application in computer vision, providing an alternative to time-consuming and crucial tasks performed by clinicians. The U-net architecture is widely used in this field and has served as the foundation for newer architectures like Swin-UNETR, which incorporates Swin transformers blocks to achieve state-of-the-art results. Various researchers have proposed different implementations using diverse convolutional blocks or modifications to the shortcut path between the encoder and decoder.
However, comparing the effectiveness of these architectural choices can be challenging due to variations in preprocessing steps and pipeline optimizations adopted by different research teams to enhance their results. Hence, this study aims to compare proposed architectural modifications against a U-net baseline, using fixed hyper-parameters, to provide meaningful comparisons on the Medical Image Segmentation Decathlon datasets (MISD).
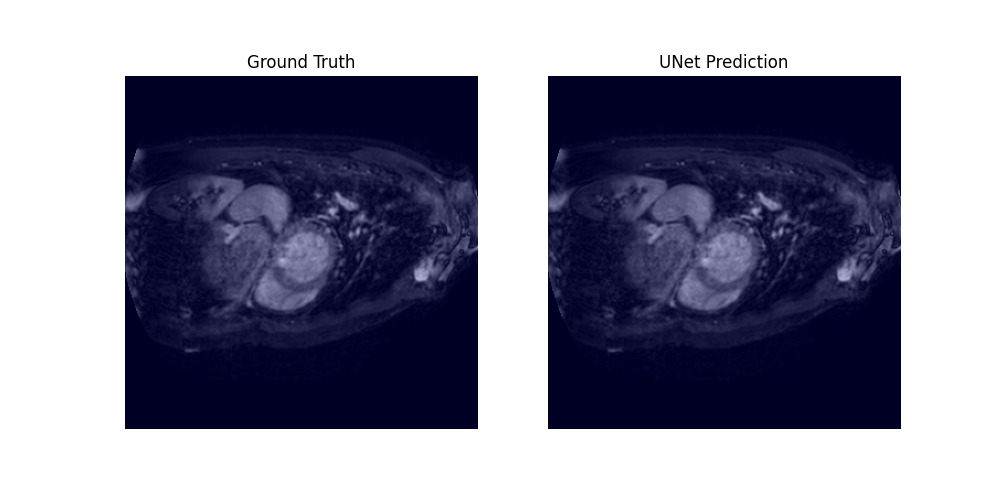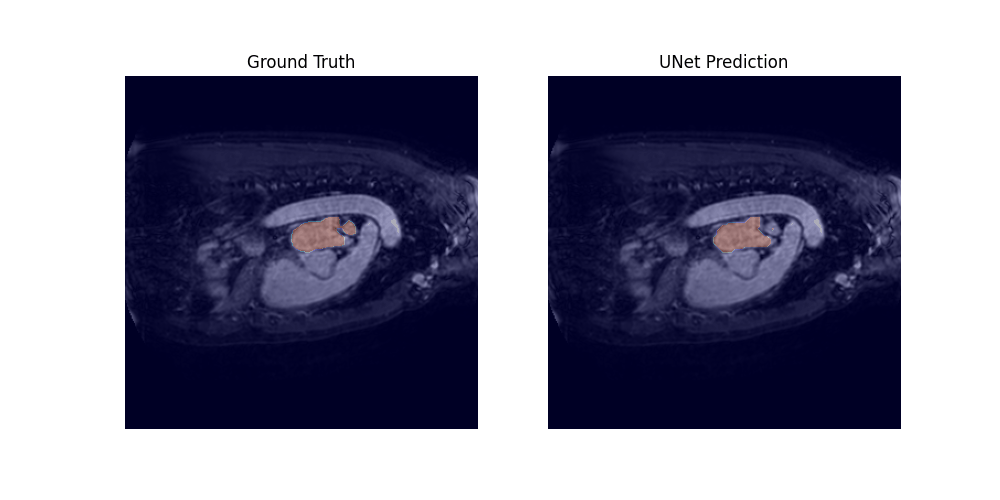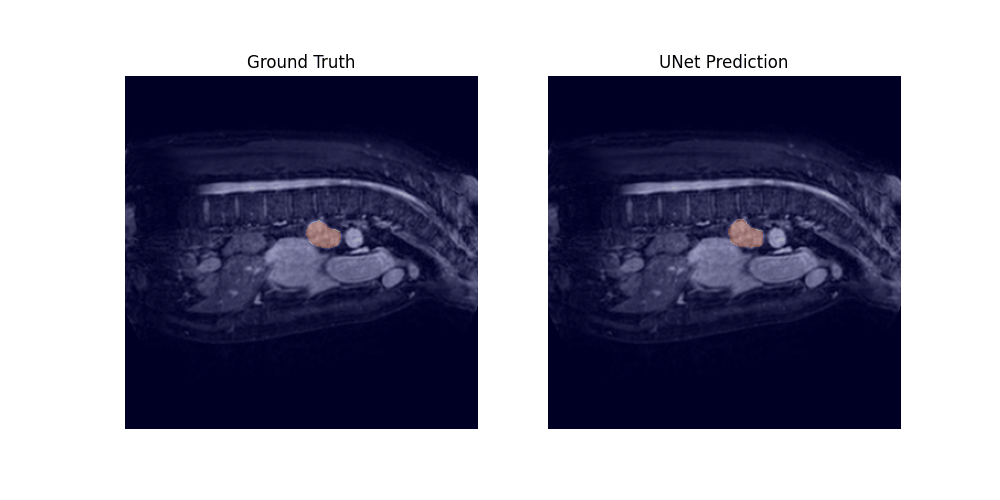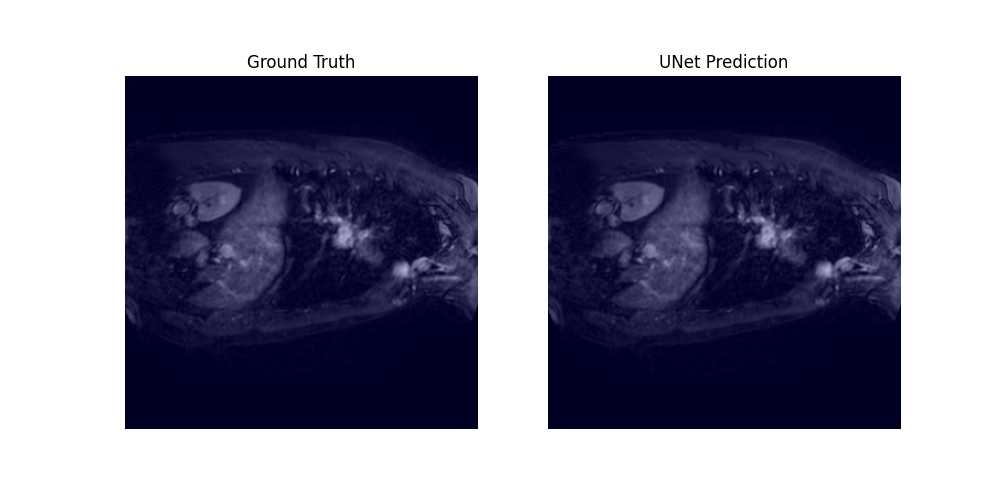
Through a 5-fold cross-validation analysis on the heart and lung datasets, we found that adding convolutional layers to the shortcut path showed promise as a design modification, although it did not reach statistical significance. Subsequently, we evaluated this modified model, along with the U-net baseline and Swin-UNETR, on all datasets, and found that it achieved the best overall performance, with an average rank of 1.59 across all tasks.
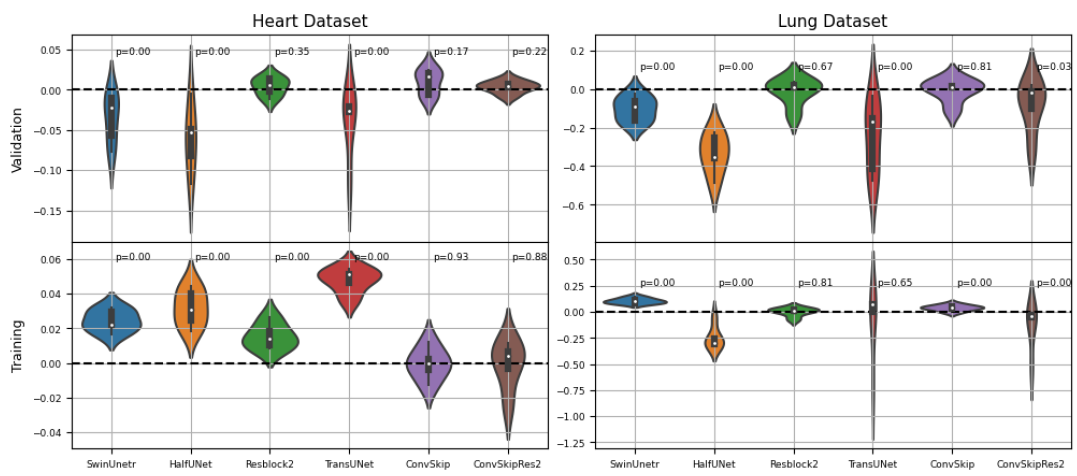


Transformers-based models did not perform as well as convolution-based ones. It is assumed that preprocessing and aggressive data augmentation may have a crucial role in the performance reported in existing literature, and further studies incorporating these factors could help bridge the gap between our study and the existing body of work.
For more details, check out the project report and the code in our repository.